Mouse: Mitochondrial and Other Useful SEquences
Version 2.0
Florian Burckhardt
MSc Biology, MSc Epidemiology
Genetic Epidemiology
Institute of Epidemiology
LMU Munich, Germany
Download:
Click conferences to get a listing of (almost) all upcoming conferences in Evolutionary Biology.
Summary:
Mouse is a user-friendly database for searching and retrieving human and great ape evolutionary sequence-data.
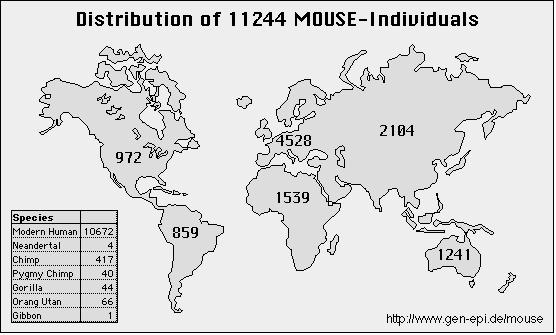
Currently, MOUSE II hosts sequences from the mitochondrial hypervariable regions I/II and chromosomal Xq13.3 sequences from humans and great apes. It contains 11,244 individuals, 5861 different HV1, 1606 different HV2 and all currently published Xq13.3 sequences (131).
With data from 95 different publications, it is the most comprehensive source for aligned mitochondrial D-loop sequences available and the only population database for Xq13.3 sequences. In addition, MOUSE II includes all relevant geographic and population-related information of the individuals, so it also serves as a highly specialised literature database for future projects.
Mouse offers a user friendly interface with online help and comes with an extensive HTML manual.
Entries can be searched for
- geographic/linguistic affiliation
- species
- sequence motif
- publication details
Search results can be summarised and saved in different formats, sequences can be manipulated within the database for further analysis.
The database is available as a free, local runtime application for PC and Mac without the need of additional programs.
Documentation available here (also included in download): http://www.gen-epi.de/mouse/docs/doc.html
"schwer ist LEicht was"
.
Changes and Version history
- 11/2001 after a little hibernation, release of MOUSE 2.0 with 4500 new individuals from 27 new publications, minor interface adjustments
- 3/2000 Mouse 1.01 for Mac and PC
- at HV2 sequence screen, HV1 sequences were saved instead of HV2 sequences
- motif search did not work because of access privilege problems
- typo corrected for Ngobe Indians (thanx to Bernal)
- "Mousehole" for faster quit added at exit sequence
- changed font to "Arial" and "New Courier" for easier PC compatibility (Yes, Arial can't live up to Helvetica. sorry Mac users)
- 2/2000 Mouse 1.0 for Mac released
- 11/1999 prerelease of Mouse for beta testing
- 4/1999 - 10/1999 working on various predecessors of Mouse, among them HvrBase, gathering user input, rebuilding codebase

1 Anderson et al.1981. Nature 290: 457-465.
page last updated Mar 14 2000




